Working Without a Diagnosis: Clinical Antimicrobial Resistance
Posted in Blog
Eva Rest, Grant Rosensteel, Emily Shambaugh, Emma Weimer
Extremely resistant pathogens, like resistant strains of Staphylococcus aureus and Clostridium difficile, are no longer susceptible to most, if any, antibiotics. C. difficile has had more response to treatment by fecal transplant or other non-antibiotic treatment measures. Overuse of systemic, broad-spectrum antibiotics and use of antibiotics to treat non-microbial or non-treatable pathogens presents significant opportunities for development of resistance in clinical settings. At this point, resistance is widespread and prevents efficient use of even “last resort” antibiotics.
Ideally, when a patient arrives at a hospital or clinic with an infectious condition, the provider could test the patient to identify the pathogen causing infection and determine susceptibility, essentially matching an effective antimicrobial treatment with the specific pathogen. In reality, diagnostic measures are not instantaneous and can take several days to produce results usable by the provider. Location or funding of a hospital can also impact access to and speed of diagnostics for infectious diseases. Patients arriving at the emergency room, immediately or soon thereafter, are often given broad-spectrum antibiotics to start addressing possible infectious conditions and prevent development of sepsis. This contributes to overuse of antibiotics for patients who may ultimately be diagnosed with a viral infection, a non-infectious condition, or a bacterial infection resistant to the prescribed broad-spectrum antibiotic. Making diagnostic tools and susceptibility analysis faster and more accurate will improve treatment, making it more patient-specific. Patients who were initially placed on broad-spectrum antibiotics should be de-escalated to narrower-spectrum antibiotics that fit their infection, or possibly taken off antibiotics entirely if the pathogen is extremely resistant. To do this, we need diagnostic tools that can produce results quickly and inform treatment choices at the point of care. Community-wide diagnostic tools, including antibiograms, can track resistance and identify geographic or hospital-specific differences in resistance to make antibiotic choices more effective.
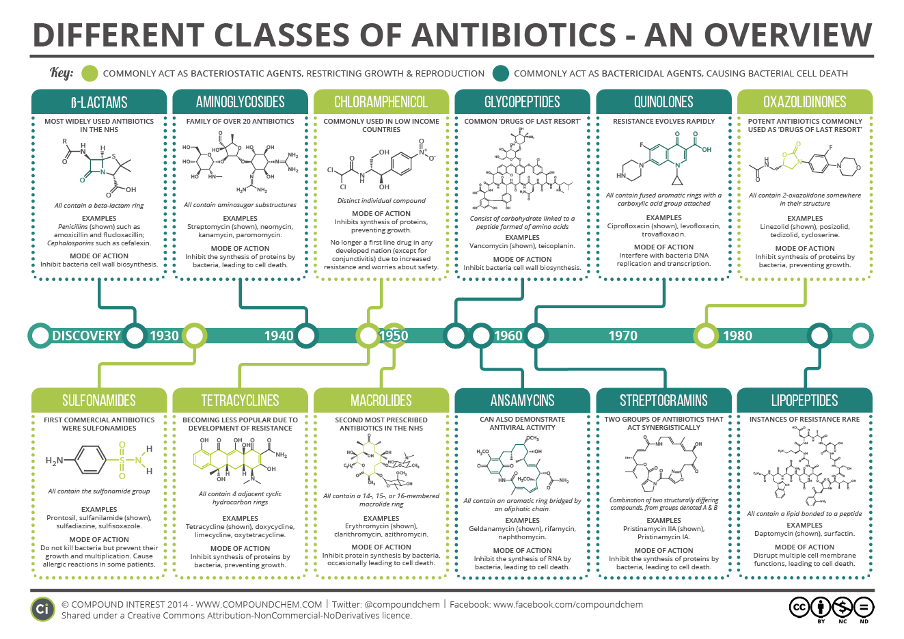
Using Antibiograms to Address Antibiotic Stewardship
On an individual level, samples from a patient likely infected with a bacterial pathogen are tested against different antimicrobial agents and doses. Susceptibility to an antibiotic is identified if the bacteria is unable to grow when exposed to the antibiotic in a lab. The minimum dose of antibiotic that prevents growth of the bacterial pathogen can be used to treat the infected patient. Antibiotics are typically effective against all or almost all target bacterial pathogens when first introduced. Over time, however, continued use of antibiotics can cause the bacterial pathogen to become resistant to the antibiotic. Resistance may start with increased doses of antibiotics required to kill the bacteria, but ultimately may render the antibiotic completely ineffective for treatment of the resistant bacterial pathogen. Antimicrobial susceptibility testing (AST) is used to evaluate which antimicrobial agents remain effective at killing the pathogen and if the pathogen has developed resistance to specific antimicrobial agents. An antibiogram is an aggregation of many AST results, compiling the data into a local or regional summary of antimicrobial resistance. Antibiograms display susceptibility and resistance data at hospital, city, or region-specific levels, whereas AST results are at the level of individual patients.
Greater access to antibiograms has the potential to help bridge the gap in time between sampling of the patient and the receipt of lab results. AST that occurs between these steps traditionally requires a 48- to 72-hour turn-around. Unfortunately, this window of time can also be critical for some antimicrobial therapeutic interventions. Within healthcare settings, antibiograms are able to provide information about the local susceptibilities of certain pathogens to a range of antimicrobial drugs. Healthcare providers can use antibiograms to avoid prescribing drugs likely to be ineffective as a result of antimicrobial resistance. Moreover, this data can be accessed before diagnostic test results are received by the provider, allowing them to prescribe a drug more likely to eliminate the infection.
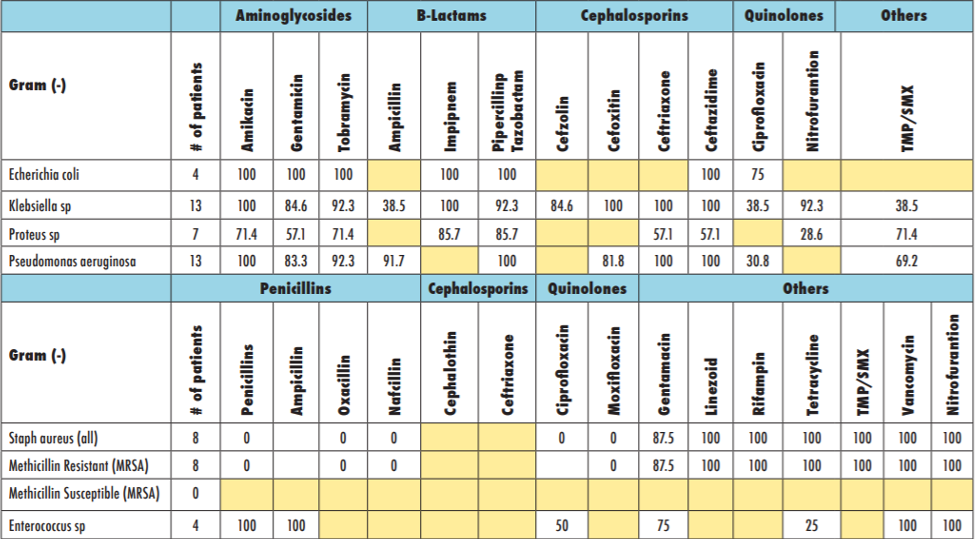
Individual hospitals and healthcare providers can contribute data to antibiograms. Additionally, community antibiograms aggregated at a city or regional level can encompass data from smaller health centers and clinics. In lieu of faster and more effective diagnostic tools within clinical settings to identify microbes and their respective susceptibilities, antibiograms can be thought of as an effective substitute. As such, their presence and use should be commonplace throughout healthcare settings in the United States to improve antimicrobial stewardship as better diagnostic processes are being developed.
Due to expense and initial complexity of setup, provider access to antibiograms is not guaranteed in healthcare settings throughout the United States. As a result of this phenomenon, Congress could incentivize health care settings, including hospitals and local health centers and clinics, to develop, continually contribute to, and utilize antibiograms. This could be implemented via an incentivizing tax break awarded annually to hospitals and health centers. This tax break would be awarded first upon verification of either a health center’s development of an independent antibiogram or their access to an existing local antibiogram. Second, verification would be required of at least three contributions to the health center’s respective antibiogram per month to ensure that data is being kept up-to-date for effective utilization. These verification processes would ensure that a hospital or health center maintains access and consistently contributes to antibiograms.
Utilizing Diagnostic Tools and Antibiograms to Enhance AMR Surveillance
The benefits of embracing diagnostic tools and antibiograms extends beyond improving the immediate point of care, but also translates into increasing antimicrobial surveillance at a large scale. The data produced by antibiograms or diagnostics on pathological agents and antimicrobial susceptibility could be collected, archived, and shared to inform the landscape of antimicrobial resistance at local, regional, and national levels. While the CDC does track antimicrobial resistance through laboratory networks, they lack the fine scale point of care surveillance necessary to combat this problem in a comprehensive manner. Without such a surveillance system, our public health providers and policy makers are at a unique disadvantage when it comes to determining proper avenues of care and when creating policies for antimicrobial use and stewardship.
In order to utilize diagnostic and antibiogram data as a surveillance mechanism, a unified and accessible reporting system would need to be established and implemented. Such a system could mirror an existing diagnostic surveillance system, like the National Respiratory and Enteric Virus Surveillance System (NREVSS) used for influenza surveillance in the U.S. In the NREVSS, the results of influenza samples from clinical and public health laboratories are reported to the CDC weekly and are displayed online for anyone to view. Antibiograms, useful for local antibiotic susceptibilities, could also be collected and aggregated to inform susceptibility surveillance. Implementing a system for antimicrobial resistance that takes into account diagnostic data and local antibiogram data would greatly advance our understanding of the antimicrobial landscape in the United States, which in turn helps advance data-driven solutions to combating AMR.
Beyond Antibiograms: Investments that could Save the Miracle of Antibiotics
While a clinician may be well-versed and enthusiastic about antimicrobial stewardship, by waiting for diagnoses before prescribing treatment, they could risk a patient’s chances of recovery. So, the clinician makes a game-time decision, without knowing lab results – also known as an empiric antibiotic therapy. Ideally, rapid diagnostics would occur in 8 hours, the length of a hospital shift, or less. Antibiograms have the capacity to shorten this period of clinical uncertainty. However, developments in AST research may offer more effective, long-term solutions. Research and development centered on modernizing AST has landed on numerous promising avenues for exploration. Automated AST, automatic disc dispensing, new ways of monitoring disc zones, detection of protein markers of resistance, and molecular detection; are all emerging technologies with promise.
Rapid diagnostics and data collection are crucial technologies for containing antibiotic resistance. Further investment into these technologies by public, private, and academic bodies have the power to save the miracle of antibiotics. With so much to gain and contribute through these investments, one must look critically at budget-making choices: are there trade-offs of investing in shorter-term versus longer-term solutions to limitations in current diagnostic technology? How can health officials restructure surveillance technologies to maximize the positive impact of improvements in diagnostics.
Resources
Bartlett JG, Gilbert DN, Spellberg B. Seven ways to preserve the miracle of antibiotics. Clinical Infectious Diseases. 2013;56(10):1445-1450. https://doi.org/10.1093/cid/cit070
Gauthier TP and Justo JA. Five important things to know about hospital antibiograms. ID Stewardship website. Updated April 2018. Accessed November 2020. https://www.idstewardship.com/five-important-things-know-hospital-antibiograms/
Minnesota Department of Health: Infectious Disease Epidemiology, Prevention, and Control Division. About antibiograms (antimicrobial susceptibilities of selected pathogens). Updated January 2015. Accessed November 2020. https://www.health.state.mn.us/diseases/antibioticresistance/abx/antibiograms.pdf
Morales A, Campos M, Juarez JM et al. A decision support system for antibiotic prescription based on local cumulative antibiograms. Journal of Biomedical Informatics. 2018;84:114-122. https://doi.org/10.1016/j.jbi.2018.07.003
Okeke IN, Peeling RW, Goosens H et al. Diagnostics as essential tools for containing antibacterial resistance. Drug Resistance Updates. 2011;14(2):95-106. https://doi.org/10.1016/j.drup.2011.02.002
Van Belkum A, Burnham CD, Rossen JA et al. Innovative and rapid antimicrobial susceptibility testing systems. Nature Reviews Microbiology. 2020;18:299-311. https://doi.org/10.1038/s41579-020-0327-x
